Nicholas (Nick) Konz

Email: nicholas (dot) konz (at) duke (dot) edu
Bluesky 🦋: @nickkonz.bsky.social
I’m an incoming postdoctoral researcher at the UNITES Lab at UNC Chapel Hill CS, where I’ll be working under Prof. Tianlong Chen starting in early 2026. My research will focus on agentic and multimodal AI for healthcare, as well as AI for science including protein and genomic language modeling. I recently completed my Ph.D. in machine learning at Duke University under Maciej Mazurowski, where my research focused on deep learning and computer vision for medical image analysis, with an emphasis on generative models, domain adaptation, and generalization analysis.
I’m also interested in how foundational deep learning concepts–such as generalization and image distribution distance metrics–need to be adapted for specialized domains like medical imaging, and how intrinsic manifold properties of training data govern model learning and generalization.
Previously, I worked as a research intern at PNNL in AI interpretability, and earned my undergraduate degree at UNC, double-majoring in physics and mathematics, where I conducted research on statistical techniques for astronomy under Dan Reichart.
To learn more about my research, check out my full list of research topics and papers.
news
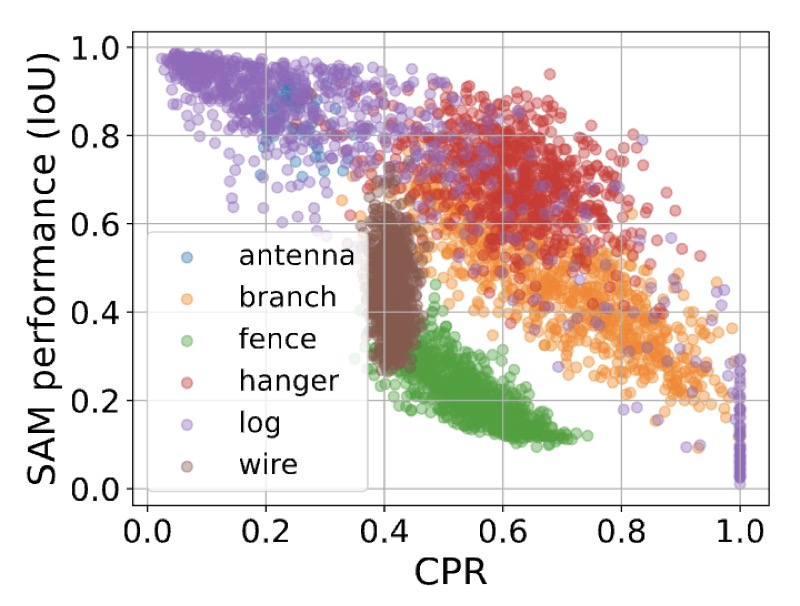
| Nov 9, 2025 | Our paper on modeling how foundation models like SAM struggle with segmenting unusual objects has been accepted to WACV 2026! |
|---|---|
| Nov 6, 2025 | I’ve successfully defended my PhD! Thank you to my advisor and committee members, and everyone in my life who has supported me along the way. |
| Oct 30, 2025 | I’m very excited to announce that I will be joining the UNITES Lab at UNC Chapel Hill as a Postdoctoral Researcher under Prof. Tianlong Chen starting in early 2026! My research will focus on multimodal and agentic AI for healthcare and science. I’m looking forward to this new chapter and the opportunity to work with this incredibly talented group! |
| Oct 27, 2025 | Our paper, “Accelerating Volumetric Medical Image Annotation via Short-Long Memory SAM 2” (preprint here) has been accepted for publication in IEEE Transactions on Medical Imaging (TMI)! |
| Oct 6, 2025 | I’m honored to have been selected to be an Area Chair for MIDL 2026! Looking forward to contributing to this fantastic conference. |
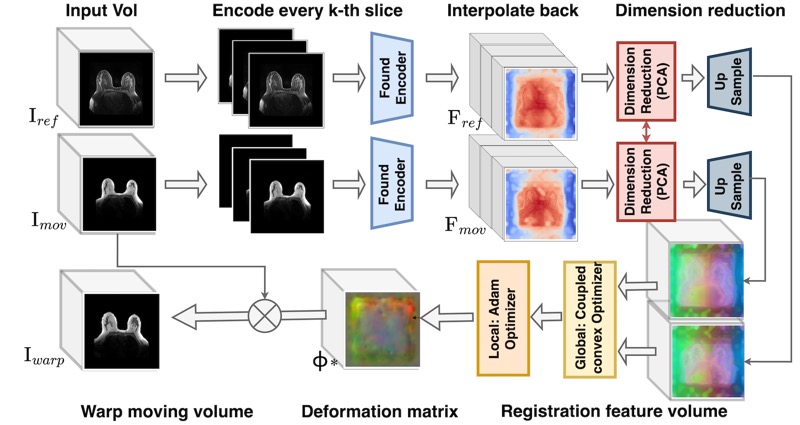
| Sep 24, 2025 | Our paper, “Are Vision Foundation Models Ready for Out-of-the-Box Medical Image Registration?” (link here), has received the best paper award at the Deep-Brea3th Workshop at MICCAI 2025! |
| Aug 12, 2025 | Our paper, “Are Vision Foundation Models Ready for Out-of-the-Box Medical Image Registration?” (link here), has been accepted and selected for an oral presentation at the Deep-Brea3th Workshop at MICCAI 2025! |
| Jun 18, 2025 | Our paper, “SegmentAnyMuscle: A universal muscle segmentation model across different locations in MRI”, has been released on the arXiv! |
| Jun 15, 2025 | Our paper, “Are Vision Foundation Models Ready for Out-of-the-Box Medical Image Registration?”, has been released on the arXiv! |
| Jun 13, 2025 | Our paper, “MRI-CORE: A Foundation Model for Magnetic Resonance Imaging”, has been released on the arXiv! |