Research Areas
My research topics of interest, past and present.
Topic List
- Topic List
- Intrinsic Properties of Image Data Manifolds and their Effects on Neural Network Generalization
- Image Distribution Similarity Metrics and Generative Models
- Vision Foundation Models for Segmentation and Beyond
- Neural Network Interpretability and Explainability
- Domain Adaptation and Analysis
- Anomaly Detection and Localization
- Misc. Breast Imaging Analysis
Intrinsic Properties of Image Data Manifolds and their Effects on Neural Network Generalization
Research exploring how geometric and statistical properties of image datasets, such as intrinsic dimension and label sharpness, influence neural network generalization across different imaging domains, particularly in how these differ between natural and medical imaging.
2024
-
 Pre-processing and Compression: Understanding Hidden Representation Refinement Across Imaging Domains via Intrinsic DimensionNicholas Konz, and Maciej A. MazurowskiSciForDL @ NeurIPS, 2024
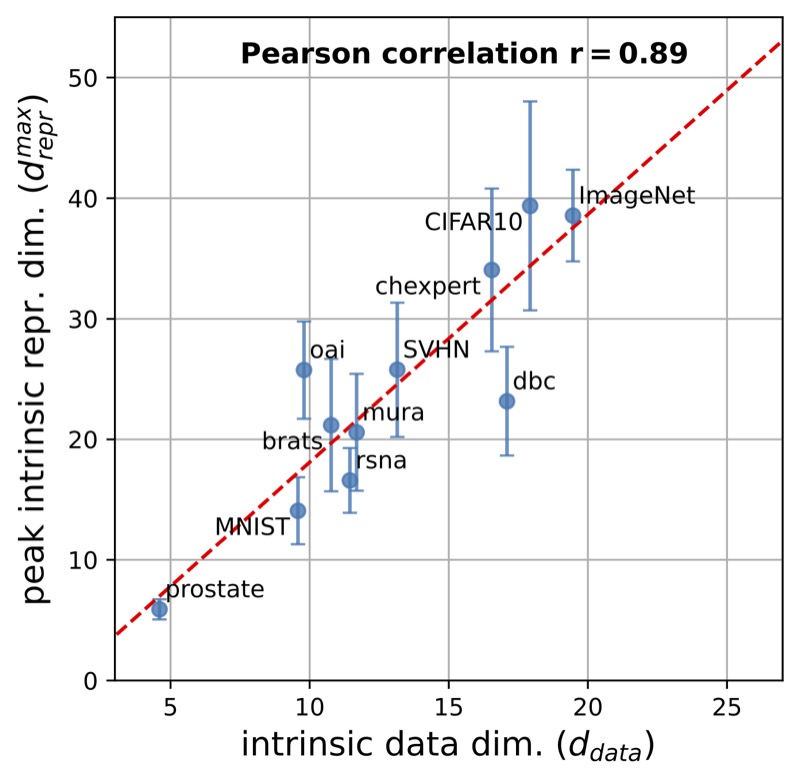
Pre-processing and Compression: Understanding Hidden Representation Refinement Across Imaging Domains via Intrinsic DimensionNicholas Konz, and Maciej A. MazurowskiSciForDL @ NeurIPS, 2024In recent years, there has been interest in how geometric properties such as intrinsic dimension (ID) of a neural network’s hidden representations change through its layers, and how such properties are predictive of important model behavior such as generalization ability. However, evidence has begun to emerge that such behavior can change significantly depending on the domain of the network’s training data, such as natural versus medical images. Here, we further this inquiry by exploring how the ID of a network’s learned representations changes through its layers, in essence, characterizing how the network successively refines the information content of input data to be used for predictions. Analyzing eleven natural and medical image datasets across six network architectures, we find that how ID changes through the network differs noticeably between natural and medical image models. Specifically, medical image models peak in representation ID earlier in the network, implying a difference in the image features and their abstractness that are typically used for downstream tasks in these domains. Additionally, we discover a strong correlation of this peak representation ID with the ID of the data in its input space, implying that the intrinsic information content of a model’s learned representations is guided by that of the data it was trained on. Overall, our findings emphasize notable discrepancies in network behavior between natural and non-natural imaging domains regarding hidden representation information content, and provide further insights into how a network’s learned features are shaped by its training data.
@article{konz2024pre, title = {Pre-processing and Compression: Understanding Hidden Representation Refinement Across Imaging Domains via Intrinsic Dimension}, author = {Konz, Nicholas and Mazurowski, Maciej A.}, journal = {SciForDL @ NeurIPS}, year = {2024}, bibtex_show = true, intrinsicproperties = {true} } -
 The Effect of Intrinsic Dataset Properties on Generalization: Unraveling Learning Differences Between Natural and Medical ImagesNicholas Konz, and Maciej A. MazurowskiICLR, 2024
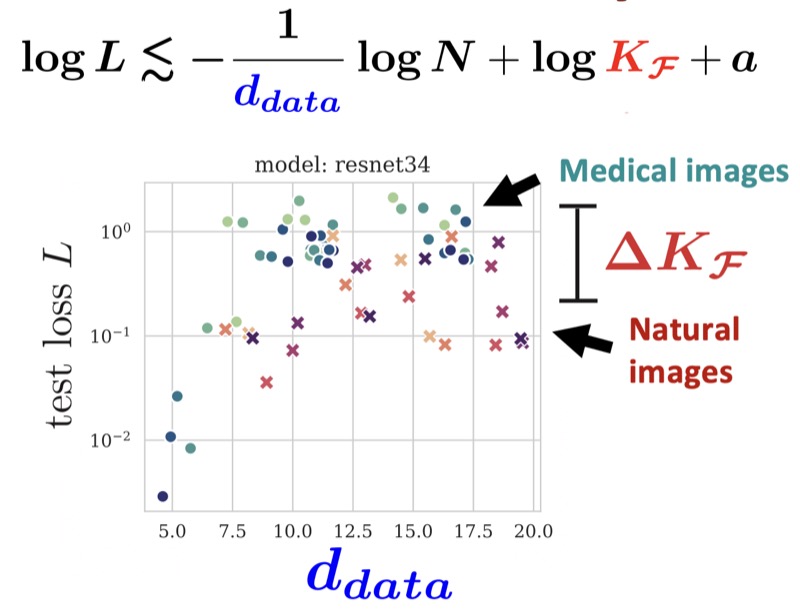
The Effect of Intrinsic Dataset Properties on Generalization: Unraveling Learning Differences Between Natural and Medical ImagesNicholas Konz, and Maciej A. MazurowskiICLR, 2024This paper investigates discrepancies in how neural networks learn from different imaging domains, which are commonly overlooked when adopting computer vision techniques from the domain of natural images to other specialized domains such as medical images. Recent works have found that the generalization error of a trained network typically increases with the intrinsic dimension (d_data) of its training set. Yet, the steepness of this relationship varies significantly between medical (radiological) and natural imaging domains, with no existing theoretical explanation. We address this gap in knowledge by establishing and empirically validating a generalization scaling law with respect to d_data, and propose that the substantial scaling discrepancy between the two considered domains may be at least partially attributed to the higher intrinsic “label sharpness” (K_F) of medical imaging datasets, a metric which we propose. Next, we demonstrate an additional benefit of measuring the label sharpness of a training set: it is negatively correlated with the trained model’s adversarial robustness, which notably leads to models for medical images having a substantially higher vulnerability to adversarial attack. Finally, we extend our d_data formalism to the related metric of learned representation intrinsic dimension (d_repr), derive a generalization scaling law with respect to d_repr, and show that d_data serves as an upper bound for d_repr. Our theoretical results are supported by thorough experiments with six models and eleven natural and medical imaging datasets over a range of training set sizes. Our findings offer insights into the influence of intrinsic dataset properties on generalization, representation learning, and robustness in deep neural networks. Code link: https://github.com/mazurowski-lab/intrinsic-properties.
@article{konz2024effect, title = {The Effect of Intrinsic Dataset Properties on Generalization: Unraveling Learning Differences Between Natural and Medical Images}, author = {Konz, Nicholas and Mazurowski, Maciej A.}, journal = {ICLR}, year = {2024}, bibtex_show = true, intrinsicproperties = {true} }
2022
-
 The Intrinsic Manifolds of Radiological Images and their Role in Deep LearningNicholas Konz, Hanxue Gu, Haoyu Dong, and 1 more authorMICCAI, 2022
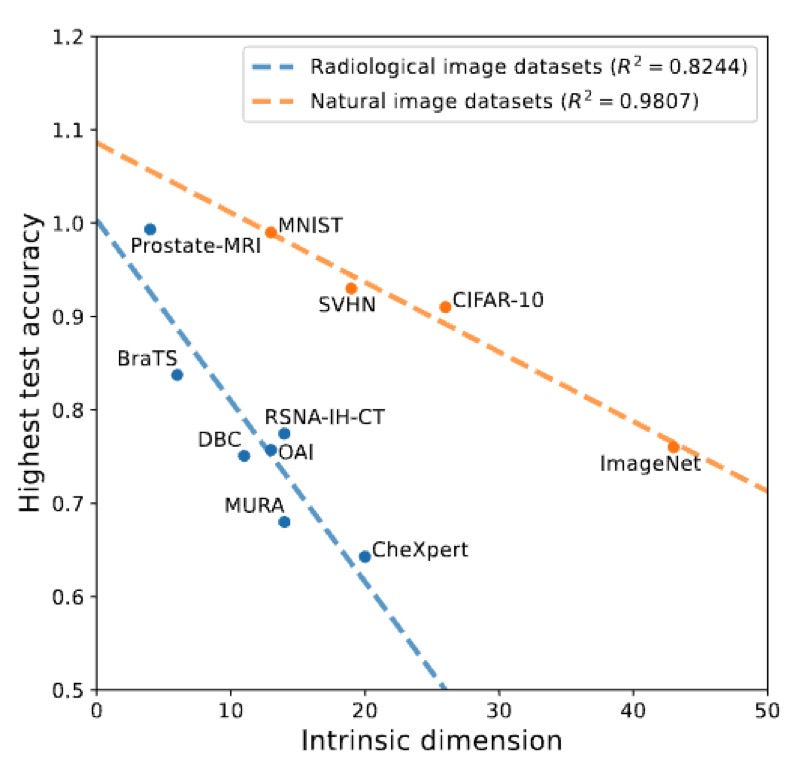
The Intrinsic Manifolds of Radiological Images and their Role in Deep LearningNicholas Konz, Hanxue Gu, Haoyu Dong, and 1 more authorMICCAI, 2022The manifold hypothesis is a core mechanism behind the success of deep learning, so understanding the intrinsic manifold structure of image data is central to studying how neural networks learn from the data. Intrinsic dataset manifolds and their relationship to learning difficulty have recently begun to be studied for the common domain of natural images, but little such research has been attempted for radiological images. We address this here. First, we compare the intrinsic manifold dimensionality of radiological and natural images. We also investigate the relationship between intrinsic dimensionality and generalization ability over a wide range of datasets. Our analysis shows that natural image datasets generally have a higher number of intrinsic dimensions than radiological images. However, the relationship between generalization ability and intrinsic dimensionality is much stronger for medical images, which could be explained as radiological images having intrinsic features that are more difficult to learn. These results give a more principled underpinning for the intuition that radiological images can be more challenging to apply deep learning to than natural image datasets common to machine learning research. We believe rather than directly applying models developed for natural images to the radiological imaging domain, more care should be taken to developing architectures and algorithms that are more tailored to the specific characteristics of this domain. The research shown in our paper, demonstrating these characteristics and the differences from natural images, is an important first step in this direction.
@article{konz2022intrinsic, title = {The Intrinsic Manifolds of Radiological Images and their Role in Deep Learning}, author = {Konz, Nicholas and Gu, Hanxue and Dong, Haoyu and Mazurowski, Maciej A.}, journal = {MICCAI}, year = {2022}, pages = {684--694}, organization = {Springer}, bibtex_show = true, intrinsicproperties = {true} }
Image Distribution Similarity Metrics and Generative Models
Development of novel perceptual metrics and generative models for medical imaging, including controllable image generation methods and specialized distance measures that better capture anatomical features than traditional computer vision metrics.
2026
-
 Fréchet Radiomic Distance (FRD): A Versatile Metric for Comparing Medical Imaging DatasetsNicholas Konz*, Richard Osuala*, Preeti Verma, and 16 more authorsMedical Image Analysis, 2026
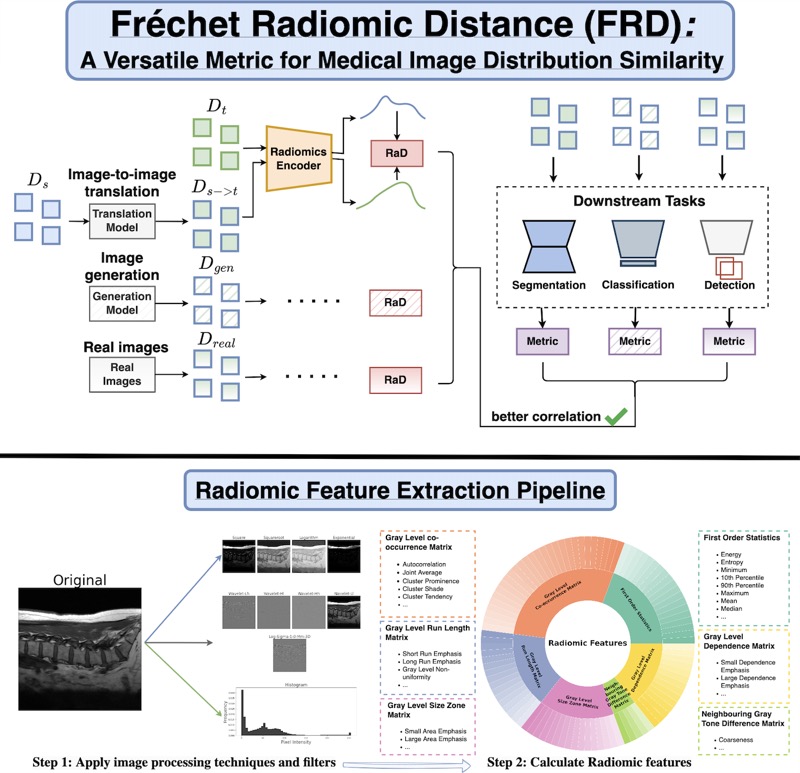
Fréchet Radiomic Distance (FRD): A Versatile Metric for Comparing Medical Imaging DatasetsNicholas Konz*, Richard Osuala*, Preeti Verma, and 16 more authorsMedical Image Analysis, 2026Determining whether two sets of images belong to the same or different distributions or domains is a crucial task in modern medical image analysis and deep learning; for example, to evaluate the output quality of image generative models. Currently, metrics used for this task either rely on the (potentially biased) choice of some downstream task, such as segmentation, or adopt task-independent perceptual metrics (\eg, Fréchet Inception Distance/FID) from natural imaging, which we show insufficiently capture anatomical features. To this end, we introduce a new perceptual metric tailored for medical images, FRD (Fréchet Radiomic Distance), which utilizes standardized, clinically meaningful, and interpretable image features. We show that FRD is superior to other image distribution metrics for a range of medical imaging applications, including out-of-domain (OOD) detection, the evaluation of image-to-image translation (by correlating more with downstream task performance as well as anatomical consistency and realism), and the evaluation of unconditional image generation. Moreover, FRD offers additional benefits such as stability and computational efficiency at low sample sizes, sensitivity to image corruptions and adversarial attacks, feature interpretability, and correlation with radiologist-perceived image quality. Additionally, we address key gaps in the literature by presenting an extensive framework for the multifaceted evaluation of image similarity metrics in medical imaging—including the first large-scale comparative study of generative models for medical image translation—and release an accessible codebase to facilitate future research. Our results are supported by thorough experiments spanning a variety of datasets, modalities, and downstream tasks, highlighting the broad potential of FRD for medical image analysis.
@article{konz2025frechetradiomicdistancefrd, title = {Fr\'echet Radiomic Distance (FRD): A Versatile Metric for Comparing Medical Imaging Datasets}, author = {Konz*, Nicholas and Osuala*, Richard and Verma, Preeti and Chen, Yuwen and Gu, Hanxue and Dong, Haoyu and Chen, Yaqian and Marshall, Andrew and Garrucho, Lidia and Kushibar, Kaisar and Lang, Daniel M. and Kim, Gene S. and Grimm, Lars J. and Lewin, John M. and Duncan, James S. and Schnabel, Julia A. and Diaz, Oliver and Lekadir, Karim and Mazurowski, Maciej A.}, year = {2026}, journal = {Medical Image Analysis}, pages = {103943}, eprint = {2412.01496}, archiveprefix = {arXiv}, primaryclass = {cs.CV}, url = {https://arxiv.org/abs/2412.01496}, bibtex_show = true, generativemodels = {true} }
2025
-
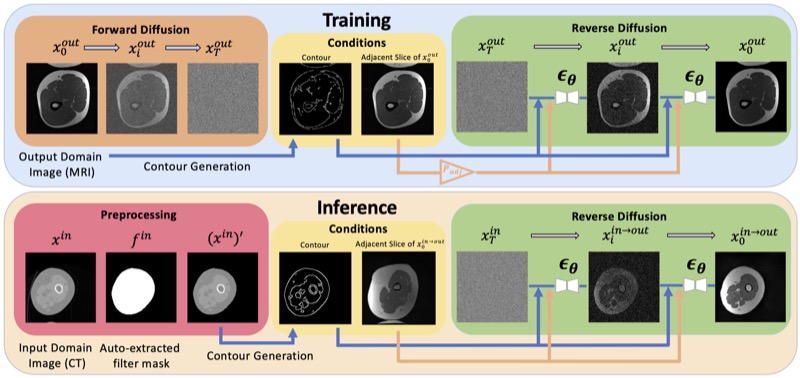 ContourDiff: Unpaired Image-to-Image Translation with Structural Consistency for Medical ImagingYuwen Chen, Nicholas Konz, Hanxue Gu, and 5 more authorsMachine Learning for Biomedical Imaging (MELBA), 2025
ContourDiff: Unpaired Image-to-Image Translation with Structural Consistency for Medical ImagingYuwen Chen, Nicholas Konz, Hanxue Gu, and 5 more authorsMachine Learning for Biomedical Imaging (MELBA), 2025Preserving object structure through image-to-image translation is crucial, particularly in applications such as medical imaging (e.g., CT-to-MRI translation), where downstream clinical and machine learning applications will often rely on such preservation. However, typical image-to-image translation algorithms prioritize perceptual quality with respect to output domain features over the preservation of anatomical structures. To address these challenges, we first introduce a novel metric to quantify the structural bias between domains which must be considered for proper translation. We then propose ContourDiff, a novel image-to-image translation algorithm that leverages domain-invariant anatomical contour representations of images to preserve the anatomical structures during translation. These contour representations are simple to extract from images, yet form precise spatial constraints on their anatomical content. ContourDiff applies an input image contour representation as a constraint at every sampling step of a diffusion model trained in the output domain, ensuring anatomical content preservation for the output image. We evaluate our method on challenging lumbar spine and hip-and-thigh CT-to-MRI translation tasks, via (1) the performance of segmentation models trained on translated images applied to real MRIs, and (2) the foreground FID and KID of translated images with respect to real MRIs. Our method outperforms other unpaired image translation methods by a significant margin across almost all metrics and scenarios. Moreover, it achieves this without the need to access any input domain information during training.
@article{chen2024contourdiff, title = {ContourDiff: Unpaired Image-to-Image Translation with Structural Consistency for Medical Imaging}, author = {Chen, Yuwen and Konz, Nicholas and Gu, Hanxue and Dong, Haoyu and Chen, Yaqian and Li, Lin and Lee, Jisoo and Mazurowski, Maciej A}, journal = {Machine Learning for Biomedical Imaging (MELBA)}, year = {2025}, bibtex_show = true, generativemodels = {true} }
2024
-
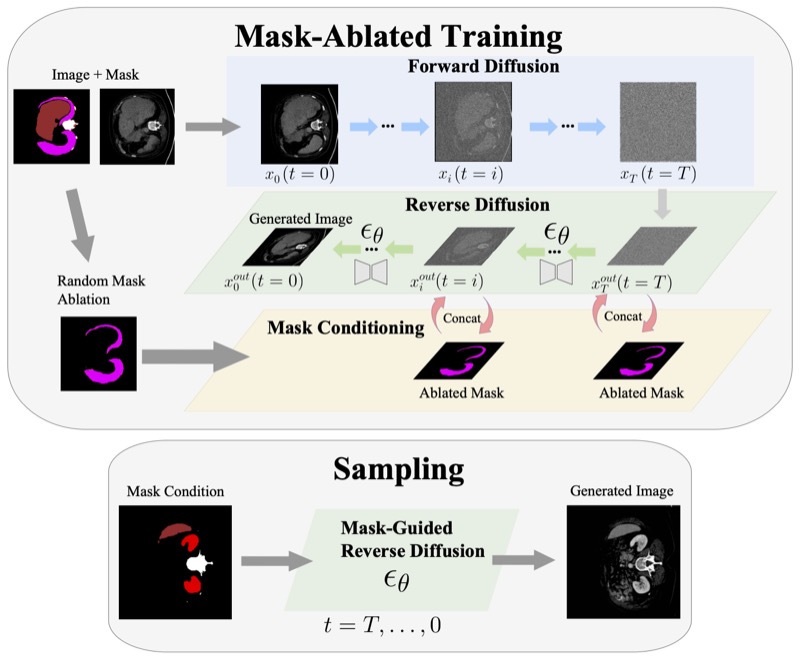 Anatomically-Controllable Medical Image Generation with Segmentation-Guided Diffusion ModelsNicholas Konz, Yuwen Chen, Haoyu Dong, and 1 more authorMICCAI, 2024
Anatomically-Controllable Medical Image Generation with Segmentation-Guided Diffusion ModelsNicholas Konz, Yuwen Chen, Haoyu Dong, and 1 more authorMICCAI, 2024Diffusion models have enabled remarkably high-quality medical image generation, yet it is challenging to enforce anatomical constraints in generated images. To this end, we propose a diffusion model-based method that supports anatomically-controllable medical image generation, by following a multi-class anatomical segmentation mask at each sampling step. We additionally introduce a random mask ablation training algorithm to enable conditioning on a selected combination of anatomical constraints while allowing flexibility in other anatomical areas. We compare our method ("SegGuidedDiff") to existing methods on breast MRI and abdominal/neck-to-pelvis CT datasets with a wide range of anatomical objects. Results show that our method reaches a new state-of-the-art in the faithfulness of generated images to input anatomical masks on both datasets, and is on par for general anatomical realism. Finally, our model also enjoys the extra benefit of being able to adjust the anatomical similarity of generated images to real images of choice through interpolation in its latent space. SegGuidedDiff has many applications, including cross-modality translation, and the generation of paired or counterfactual data. Our code is available at https://github.com/mazurowski-lab/segmentation-guided-diffusion.
@article{konz2024anatomically, title = {Anatomically-Controllable Medical Image Generation with Segmentation-Guided Diffusion Models}, author = {Konz, Nicholas and Chen, Yuwen and Dong, Haoyu and Mazurowski, Maciej A.}, journal = {MICCAI}, year = {2024}, organization = {Springer}, bibtex_show = true, generativemodels = {true} } -
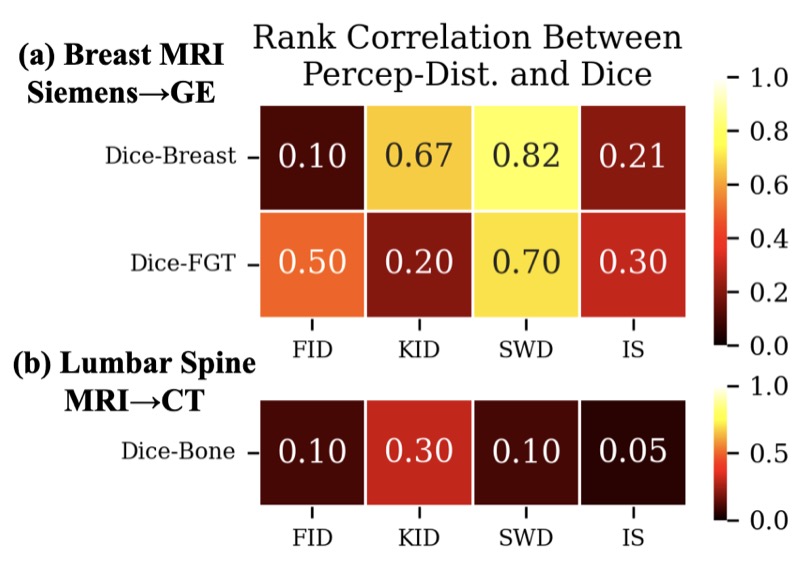 Rethinking Perceptual Metrics for Medical Image TranslationNicholas Konz, Yuwen Chen, Hanxue Gu, and 2 more authorsMIDL, 2024
Rethinking Perceptual Metrics for Medical Image TranslationNicholas Konz, Yuwen Chen, Hanxue Gu, and 2 more authorsMIDL, 2024Modern medical image translation methods use generative models for tasks such as the conversion of CT images to MRI. Evaluating these methods typically relies on some chosen downstream task in the target domain, such as segmentation. On the other hand, task-agnostic metrics are attractive, such as the network feature-based perceptual metrics (e.g., FID) that are common to image translation in general computer vision. In this paper, we investigate evaluation metrics for medical image translation on two medical image translation tasks (GE breast MRI to Siemens breast MRI and lumbar spine MRI to CT), tested on various state-of-the-art translation methods. We show that perceptual metrics do not generally correlate with segmentation metrics due to them extending poorly to the anatomical constraints of this sub-field, with FID being especially inconsistent. However, we find that the lesser-used pixel-level SWD metric may be useful for subtle intra-modality translation. Our results demonstrate the need for further research into helpful metrics for medical image translation.
@article{konz2024rethinking, title = {Rethinking Perceptual Metrics for Medical Image Translation}, author = {Konz, Nicholas and Chen, Yuwen and Gu, Hanxue and Dong, Haoyu and Mazurowski, Maciej A.}, journal = {MIDL}, year = {2024}, bibtex_show = true, generativemodels = {true} }
Vision Foundation Models for Segmentation and Beyond
Investigation of the capabilities and limitations of large-scale foundation models for medical image analysis, including enhanced architectures and performance evaluation across diverse downstream tasks.
2026
-
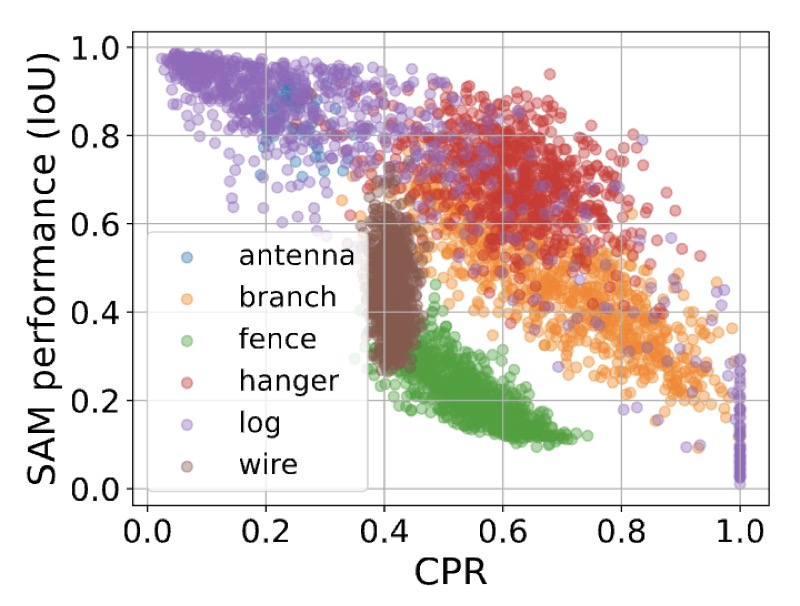 Quantifying the Limits of Segmentation Foundation Models: Modeling Challenges in Segmenting Tree-Like and Low-Contrast ObjectsYixin Zhang*, Nicholas Konz*, Kevin Kramer, and 1 more authorWACV, 2026
Quantifying the Limits of Segmentation Foundation Models: Modeling Challenges in Segmenting Tree-Like and Low-Contrast ObjectsYixin Zhang*, Nicholas Konz*, Kevin Kramer, and 1 more authorWACV, 2026Image segmentation foundation models (SFMs) like Segment Anything Model (SAM) have achieved impressive zero-shot and interactive segmentation across diverse domains. However, they struggle to segment objects with certain structures, particularly those with dense, tree-like morphology and low textural contrast from their surroundings. These failure modes are crucial for understanding the limitations of SFMs in real-world applications. To systematically study this issue, we introduce interpretable metrics quantifying object tree-likeness and textural separability. On carefully controlled synthetic experiments and real-world datasets, we show that SFM performance (e.g., SAM, SAM 2, HQ-SAM) noticeably correlates with these factors. We link these failures to "textural confusion", where models misinterpret local structure as global texture, causing over-segmentation or difficulty distinguishing objects from similar backgrounds. Notably, targeted fine-tuning fails to resolve this issue, indicating a fundamental limitation. Our study provides the first quantitative framework for modeling the behavior of SFMs on challenging structures, offering interpretable insights into their segmentation capabilities.
@article{zhang2024quantifying, title = {Quantifying the Limits of Segmentation Foundation Models: Modeling Challenges in Segmenting Tree-Like and Low-Contrast Objects}, author = {Zhang*, Yixin and Konz*, Nicholas and Kramer, Kevin and Mazurowski, Maciej A.}, journal = {WACV}, year = {2026}, bibtex_show = true, foundationmodels = {true} }
2025
-
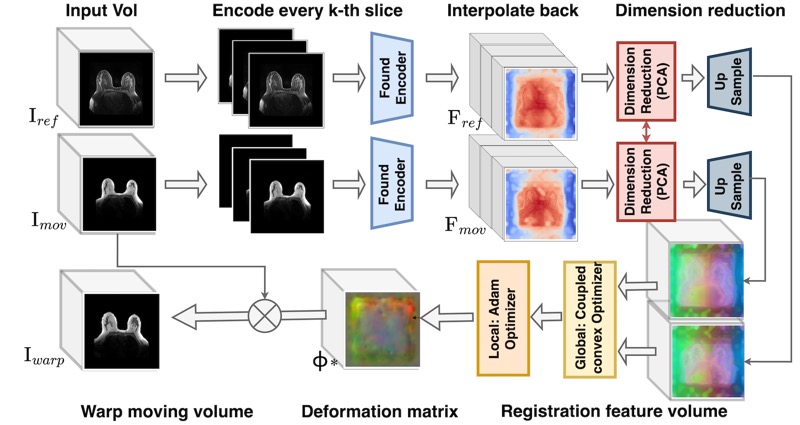 Are Vision Foundation Models Ready for Out-of-the-Box Medical Image Registration?Hanxue Gu*, Yaqian Chen*, Nicholas Konz, and 2 more authorsDeep-Breath @ MICCAI (Oral, Best Paper Award), 2025
Are Vision Foundation Models Ready for Out-of-the-Box Medical Image Registration?Hanxue Gu*, Yaqian Chen*, Nicholas Konz, and 2 more authorsDeep-Breath @ MICCAI (Oral, Best Paper Award), 2025Foundation models, pre-trained on large image datasets and capable of capturing rich feature representations, have recently shown potential for zero-shot image registration. However, their performance has mostly been tested in the context of rigid or less complex structures, such as the brain or abdominal organs, and it remains unclear whether these models can handle more challenging, deformable anatomy. Breast MRI registration is particularly difficult due to significant anatomical variation between patients, deformation caused by patient positioning, and the presence of thin and complex internal structure of fibroglandular tissue, where accurate alignment is crucial. Whether foundation model-based registration algorithms can address this level of complexity remains an open question. In this study, we provide a comprehensive evaluation of foundation model-based registration algorithms for breast MRI. We assess five pre-trained encoders, including DINO-v2, SAM, MedSAM, SSLSAM, and MedCLIP, across four key breast registration tasks that capture variations in different years and dates, sequences, modalities, and patient disease status (lesion versus no lesion). Our results show that foundation model-based algorithms such as SAM outperform traditional registration baselines for overall breast alignment, especially under large domain shifts, but struggle with capturing fine details of fibroglandular tissue. Interestingly, additional pre-training or fine-tuning on medical or breast-specific images in MedSAM and SSLSAM, does not improve registration performance and may even decrease it in some cases. Further work is needed to understand how domain-specific training influences registration and to explore targeted strategies that improve both global alignment and fine structure accuracy. We also publicly release our code at https://github.com/mazurowski-lab/Foundation-based-reg.
@article{gu2025vision, title = {Are Vision Foundation Models Ready for Out-of-the-Box Medical Image Registration?}, author = {Gu*, Hanxue and Chen*, Yaqian and Konz, Nicholas and Li, Qihang and Mazurowski, Maciej A}, journal = {Deep-Breath @ MICCAI (Oral, Best Paper Award)}, year = {2025}, bibtex_show = true, foundationmodels = {true} } -
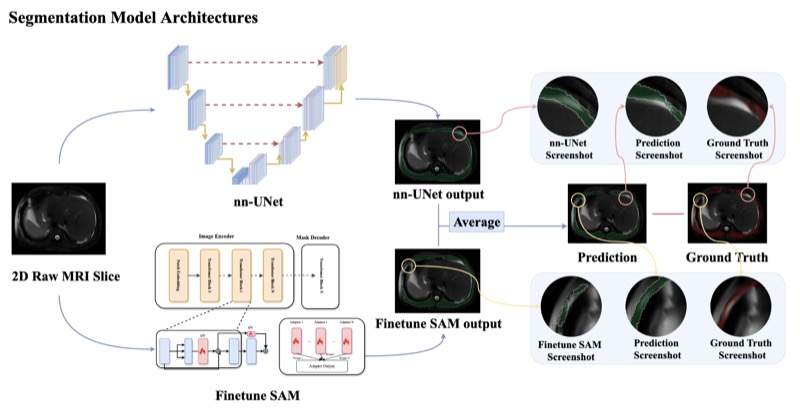 SegmentAnyMuscle: A universal muscle segmentation model across different locations in MRIRoy Colglazier, Jisoo Lee, Haoyu Dong, and 8 more authorsarXiv preprint, 2025
SegmentAnyMuscle: A universal muscle segmentation model across different locations in MRIRoy Colglazier, Jisoo Lee, Haoyu Dong, and 8 more authorsarXiv preprint, 2025The quantity and quality of muscles are increasingly recognized as important predictors of health outcomes. While MRI offers a valuable modality for such assessments, obtaining precise quantitative measurements of musculature remains challenging. This study aimed to develop a publicly available model for muscle segmentation in MRIs and demonstrate its applicability across various anatomical locations and imaging sequences. A total of 362 MRIs from 160 patients at a single tertiary center (Duke University Health System, 2016-2020) were included, with 316 MRIs from 114 patients used for model development. The model was tested on two separate sets: one with 28 MRIs representing common sequence types, achieving an average Dice Similarity Coefficient (DSC) of 88.45%, and another with 18 MRIs featuring less frequent sequences and abnormalities such as muscular atrophy, hardware, and significant noise, achieving 86.21% DSC. These results demonstrate the feasibility of a fully automated deep learning algorithm for segmenting muscles on MRI across diverse settings. The public release of this model enables consistent, reproducible research into the relationship between musculature and health.
@article{colglazier2025segmentanymuscle, title = {SegmentAnyMuscle: A universal muscle segmentation model across different locations in MRI}, author = {Colglazier, Roy and Lee, Jisoo and Dong, Haoyu and Gu, Hanxue and Chen, Yaqian and Cao, Joseph and Yildiz, Zafer and Liu, Zhonghao and Konz, Nicholas and Yang, Jichen and others}, journal = {arXiv preprint}, year = {2025}, bibtex_show = true, foundationmodels = {true} } -
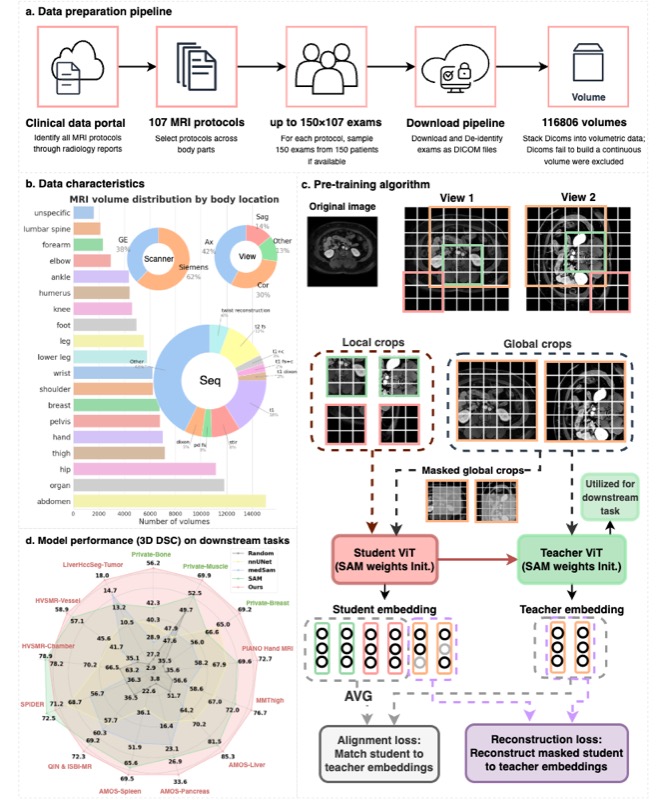 MRI-CORE: A Foundation Model for Magnetic Resonance ImagingHaoyu Dong, Yuwen Chen, Hanxue Gu, and 4 more authorsarXiv preprint, 2025
MRI-CORE: A Foundation Model for Magnetic Resonance ImagingHaoyu Dong, Yuwen Chen, Hanxue Gu, and 4 more authorsarXiv preprint, 2025The widespread use of Magnetic Resonance Imaging (MRI) in combination with deep learning shows promise for many high-impact automated diagnostic and prognostic tools. However, training new models requires large amounts of labeled data, a challenge due to high cost of precise annotations and data privacy. To address this issue, we introduce the MRI-CORE, a vision foundation model trained using more than 6 million slices from over 110 thousand MRI volumes across 18 body locations. Our experiments show notable improvements in performance over state-of-the-art methods in 13 data-restricted segmentation tasks, as well as in image classification, and zero-shot segmentation, showing the strong potential of MRI-CORE to enable data-efficient development of artificial intelligence models. We also present data on which strategies yield most useful foundation models and a novel analysis relating similarity between pre-training and downstream task data with transfer learning performance. Our model is publicly available with a permissive license.
@article{dong2025mri, title = {MRI-CORE: A Foundation Model for Magnetic Resonance Imaging}, author = {Dong, Haoyu and Chen, Yuwen and Gu, Hanxue and Konz, Nicholas and Chen, Yaqian and Li, Qihang and Mazurowski, Maciej A}, journal = {arXiv preprint}, year = {2025}, bibtex_show = true, foundationmodels = {true} } -
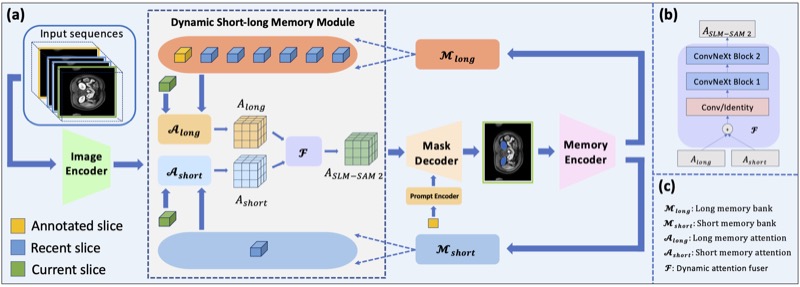 Accelerating Volumetric Medical Image Annotation via Short-Long Memory SAM 2Yuwen Chen, Zafer Yildiz, Qihang Li, and 5 more authorsIEEE Transactions on Medical Imaging, 2025
Accelerating Volumetric Medical Image Annotation via Short-Long Memory SAM 2Yuwen Chen, Zafer Yildiz, Qihang Li, and 5 more authorsIEEE Transactions on Medical Imaging, 2025Manual annotation of volumetric medical images, such as magnetic resonance imaging (MRI) and computed tomography (CT), is a labor-intensive and time-consuming process. Recent advancements in foundation models for video object segmentation, such as Segment Anything Model 2 (SAM 2), offer a potential opportunity to significantly speed up the annotation process by manually annotating one or a few slices and then propagating target masks across the entire volume. However, the performance of SAM 2 in this context varies. Our experiments show that relying on a single memory bank and attention module is prone to error propagation, particularly at boundary regions where the target is present in the previous slice but absent in the current one. To address this problem, we propose Short-Long Memory SAM 2 (SLM-SAM 2), a novel architecture that integrates distinct short-term and long-term memory banks with separate attention modules to improve segmentation accuracy. We evaluate SLM-SAM 2 on three public datasets covering organs, bones, and muscles across MRI and CT modalities. We show that the proposed method markedly outperforms the default SAM 2, achieving average Dice Similarity Coefficient improvement of 0.14 and 0.11 in the scenarios when 5 volumes and 1 volume are available for the initial adaptation, respectively. SLM-SAM 2 also exhibits stronger resistance to over-propagation, making a notable step toward more accurate automated annotation of medical images for segmentation model development.
@article{chen2025accelerating, title = {Accelerating Volumetric Medical Image Annotation via Short-Long Memory SAM 2}, author = {Chen, Yuwen and Yildiz, Zafer and Li, Qihang and Chen, Yaqian and Dong, Haoyu and Gu, Hanxue and Konz, Nicholas and Mazurowski, Maciej A}, journal = {IEEE Transactions on Medical Imaging}, year = {2025}, bibtex_show = true, foundationmodels = {true} }
2023
-
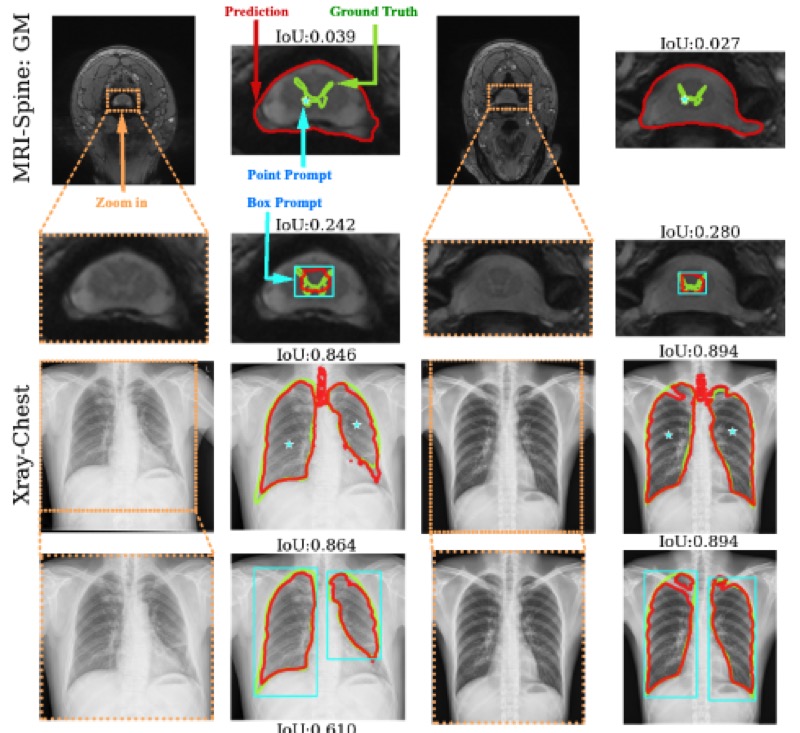 Segment anything model for medical image analysis: an experimental studyMaciej A. Mazurowski, Haoyu Dong, Hanxue Gu, and 3 more authorsMedical Image Analysis, 2023
Segment anything model for medical image analysis: an experimental studyMaciej A. Mazurowski, Haoyu Dong, Hanxue Gu, and 3 more authorsMedical Image Analysis, 2023Training segmentation models for medical images continues to be challenging due to the limited availability of data annotations. Segment Anything Model (SAM) is a foundation model that is intended to segment user-defined objects of interest in an interactive manner. While the performance on natural images is impressive, medical image domains pose their own set of challenges. Here, we perform an extensive evaluation of SAM’s ability to segment medical images on a collection of 19 medical imaging datasets from various modalities and anatomies. We report the following findings: (1) SAM’s performance based on single prompts highly varies depending on the dataset and the task, from IoU=0.1135 for spine MRI to IoU=0.8650 for hip X-ray. (2) Segmentation performance appears to be better for well-circumscribed objects with prompts with less ambiguity and poorer in various other scenarios such as the segmentation of brain tumors. (3) SAM performs notably better with box prompts than with point prompts. (4) SAM outperforms similar methods RITM, SimpleClick, and FocalClick in almost all single-point prompt settings. (5) When multiple-point prompts are provided iteratively, SAM’s performance generally improves only slightly while other methods’ performance improves to the level that surpasses SAM’s point-based performance. We also provide several illustrations for SAM’s performance on all tested datasets, iterative segmentation, and SAM’s behavior given prompt ambiguity. We conclude that SAM shows impressive zero-shot segmentation performance for certain medical imaging datasets, but moderate to poor performance for others. SAM has the potential to make a significant impact in automated medical image segmentation in medical imaging, but appropriate care needs to be applied when using it.
@article{mazurowski2023segment, title = {Segment anything model for medical image analysis: an experimental study}, author = {Mazurowski, Maciej A. and Dong, Haoyu and Gu, Hanxue and Yang, Jichen and Konz, Nicholas and Zhang, Yixin}, journal = {Medical Image Analysis}, volume = {89}, pages = {102918}, year = {2023}, publisher = {Elsevier}, bibtex_show = true, foundationmodels = {true} }
Neural Network Interpretability and Explainability
Methods for understanding what neural networks learn and how concepts are formed, including data attribution techniques and representation analysis to improve model transparency and trustworthiness, and others.
2023
-
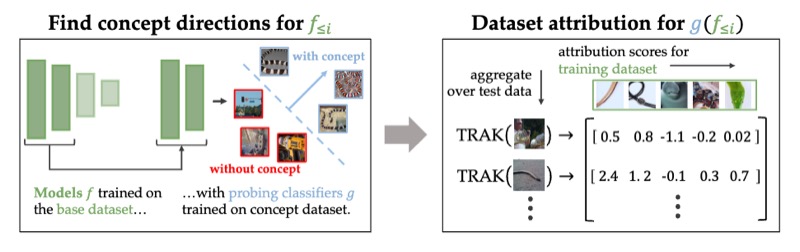 Attributing Learned Concepts in Neural Networks to Training DataNicholas Konz, Charles Godfrey, Madelyn Shapiro, and 3 more authorsATTRIB @ NeurIPS (Oral), 2023
Attributing Learned Concepts in Neural Networks to Training DataNicholas Konz, Charles Godfrey, Madelyn Shapiro, and 3 more authorsATTRIB @ NeurIPS (Oral), 2023By now there is substantial evidence that deep learning models learn certain human-interpretable features as part of their internal representations of data. As having the right (or wrong) concepts is critical to trustworthy machine learning systems, it is natural to ask which inputs from the model’s original training set were most important for learning a concept at a given layer. To answer this, we combine data attribution methods with methods for probing the concepts learned by a model. Training network and probe ensembles for two concept datasets on a range of network layers, we use the recently developed TRAK method for large-scale data attribution. We find some evidence for convergence, where removing the 10,000 top attributing images for a concept and retraining the model does not change the location of the concept in the network nor the probing sparsity of the concept. This suggests that rather than being highly dependent on a few specific examples, the features that inform the development of a concept are spread in a more diffuse manner across its exemplars, implying robustness in concept formation.
@article{konz2023attributing, title = {Attributing Learned Concepts in Neural Networks to Training Data}, author = {Konz, Nicholas and Godfrey, Charles and Shapiro, Madelyn and Tu, Jonathan and Kvinge, Henry and Brown, Davis}, journal = {ATTRIB @ NeurIPS (Oral)}, year = {2023}, bibtex_show = true, interpretability = {true} } -
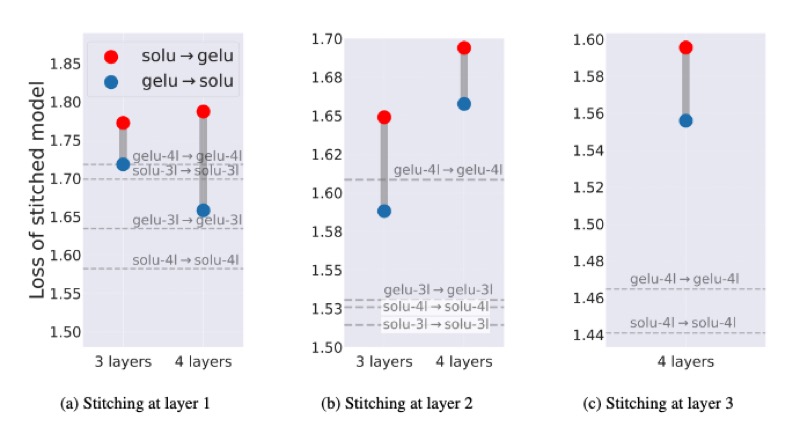 Understanding the Inner-workings of Language Models Through Representation DissimilarityDavis Brown, Charles Godfrey, Nicholas Konz, and 2 more authorsEMNLP, 2023
Understanding the Inner-workings of Language Models Through Representation DissimilarityDavis Brown, Charles Godfrey, Nicholas Konz, and 2 more authorsEMNLP, 2023As language models are applied to an increasing number of real-world applications, understanding their inner workings has become an important issue in model trust, interpretability, and transparency. In this work we show that representation dissimilarity measures, which are functions that measure the extent to which two model’s internal representations differ, can be a valuable tool for gaining insight into the mechanics of language models. Among our insights are: (i) an apparent asymmetry in the internal representations of model using SoLU and GeLU activation functions, (ii) evidence that dissimilarity measures can identify and locate generalization properties of models that are invisible via in-distribution test set performance, and (iii) new evaluations of how language model features vary as width and depth are increased. Our results suggest that dissimilarity measures are a promising set of tools for shedding light on the inner workings of language models.
@article{brown2023understanding, title = {Understanding the Inner-workings of Language Models Through Representation Dissimilarity}, author = {Brown, Davis and Godfrey, Charles and Konz, Nicholas and Tu, Jonathan and Kvinge, Henry}, journal = {EMNLP}, year = {2023}, bibtex_show = true, interpretability = {true} } -
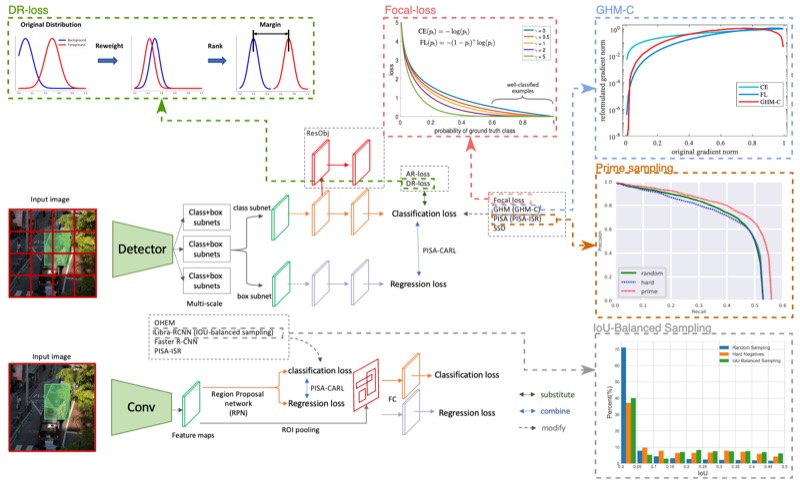 A systematic study of the foreground-background imbalance problem in deep learning for object detectionHanxue Gu, Haoyu Dong, Nicholas Konz, and 1 more authorarXiv preprint, 2023
A systematic study of the foreground-background imbalance problem in deep learning for object detectionHanxue Gu, Haoyu Dong, Nicholas Konz, and 1 more authorarXiv preprint, 2023The class imbalance problem in deep learning has been explored in several studies, but there has yet to be a systematic analysis of this phenomenon in object detection. Here, we present comprehensive analyses and experiments of the foreground-background (F-B) imbalance problem in object detection, which is very common and caused by small, infrequent objects of interest. We experimentally study the effects of different aspects of F-B imbalance (object size, number of objects, dataset size, object type) on detection performance. In addition, we also compare 9 leading methods for addressing this problem, including Faster-RCNN, SSD, OHEM, Libra-RCNN, Focal-Loss, GHM, PISA, YOLO-v3, and GFL with a range of datasets from different imaging domains. We conclude that (1) the F-B imbalance can indeed cause a significant drop in detection performance, (2) The detection performance is more affected by F-B imbalance when fewer training data are available, (3) in most cases, decreasing object size leads to larger performance drop than decreasing number of objects, given the same change in the ratio of object pixels to non-object pixels, (6) among all selected methods, Libra-RCNN and PISA demonstrate the best performance in addressing the issue of F-B imbalance. (7) When the training dataset size is large, the choice of method is not impactful (8) Soft-sampling methods, including focal-loss, GHM, and GFL, perform fairly well on average but are relatively unstable.
@article{gu2023systematic, title = {A systematic study of the foreground-background imbalance problem in deep learning for object detection}, author = {Gu, Hanxue and Dong, Haoyu and Konz, Nicholas and Mazurowski, Maciej A}, journal = {arXiv preprint}, year = {2023}, bibtex_show = true, interpretability = {true} }
Domain Adaptation and Analysis
Techniques for adapting models across different imaging domains, scanners, and acquisition parameters, addressing the challenge of domain shift that commonly affects medical ML systems in practice, as well as understanding the nature of the domain shift problem itself.
2024
-
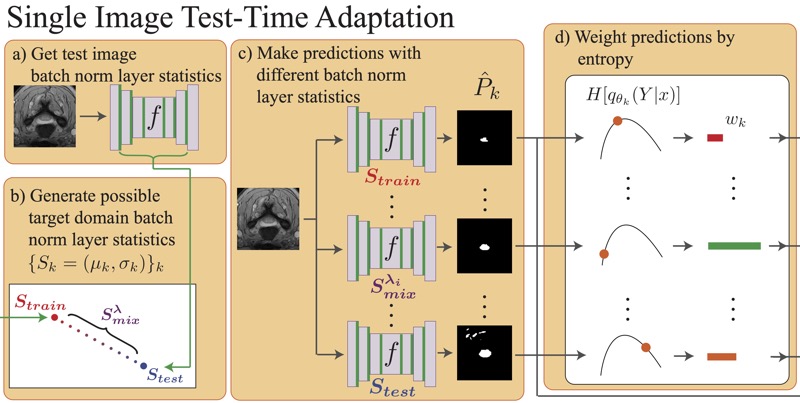 Medical Image Segmentation with InTEnt: Integrated Entropy Weighting for Single Image Test-Time AdaptationHaoyu Dong, Nicholas Konz, Hanxue Gu, and 1 more authorDEF-AI-MIA @ CVPR (Oral), 2024
Medical Image Segmentation with InTEnt: Integrated Entropy Weighting for Single Image Test-Time AdaptationHaoyu Dong, Nicholas Konz, Hanxue Gu, and 1 more authorDEF-AI-MIA @ CVPR (Oral), 2024Test-time adaptation (TTA) refers to adapting a trained model to a new domain during testing. Existing TTA techniques rely on having multiple test images from the same domain, yet this may be impractical in real-world applications such as medical imaging, where data acquisition is expensive and imaging conditions vary frequently. Here, we approach such a task, of adapting a medical image segmentation model with only a single unlabeled test image. Most TTA approaches, which directly minimize the entropy of predictions, fail to improve performance significantly in this setting, in which we also observe the choice of batch normalization (BN) layer statistics to be a highly important yet unstable factor due to only having a single test domain example. To overcome this, we propose to instead integrate over predictions made with various estimates of target domain statistics between the training and test statistics, weighted based on their entropy statistics. Our method, validated on 24 source/target domain splits across 3 medical image datasets surpasses the leading method by 2.9% Dice coefficient on average.
@article{dong2024intent, title = {Medical Image Segmentation with InTEnt: Integrated Entropy Weighting for Single Image Test-Time Adaptation}, author = {Dong, Haoyu and Konz, Nicholas and Gu, Hanxue and Mazurowski, Maciej A.}, journal = {DEF-AI-MIA @ CVPR (Oral)}, year = {2024}, bibtex_show = true, domainadaptation = {true} } -
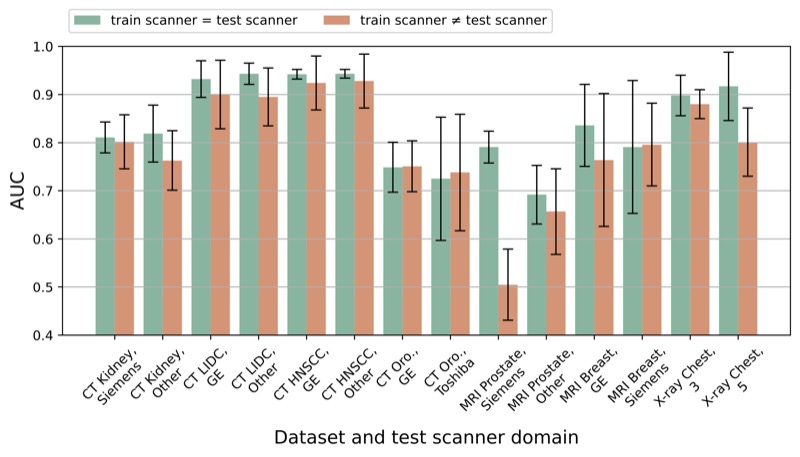 The impact of scanner domain shift on deep learning performance in medical imaging: an experimental studyBrian Guo, Darui Lu, Gregory Szumel, and 4 more authorsarXiv preprint, 2024
The impact of scanner domain shift on deep learning performance in medical imaging: an experimental studyBrian Guo, Darui Lu, Gregory Szumel, and 4 more authorsarXiv preprint, 2024Purpose: Medical images acquired using different scanners and protocols can differ substantially in their appearance. This phenomenon, scanner domain shift, can result in a drop in the performance of deep neural networks which are trained on data acquired by one scanner and tested on another. This significant practical issue is well-acknowledged, however, no systematic study of the issue is available across different modalities and diagnostic tasks. Materials and Methods: In this paper, we present a broad experimental study evaluating the impact of scanner domain shift on convolutional neural network performance for different automated diagnostic tasks. We evaluate this phenomenon in common radiological modalities, including X-ray, CT, and MRI. Results: We find that network performance on data from a different scanner is almost always worse than on same-scanner data, and we quantify the degree of performance drop across different datasets. Notably, we find that this drop is most severe for MRI, moderate for X-ray, and quite small for CT, on average, which we attribute to the standardized nature of CT acquisition systems which is not present in MRI or X-ray. We also study how injecting varying amounts of target domain data into the training set, as well as adding noise to the training data, helps with generalization. Conclusion: Our results provide extensive experimental evidence and quantification of the extent of performance drop caused by scanner domain shift in deep learning across different modalities, with the goal of guiding the future development of robust deep learning models for medical image analysis.
@article{guo2024impact, title = {The impact of scanner domain shift on deep learning performance in medical imaging: an experimental study}, author = {Guo, Brian and Lu, Darui and Szumel, Gregory and Gui, Rongze and Wang, Tingyu and Konz, Nicholas and Mazurowski, Maciej A}, journal = {arXiv preprint}, year = {2024}, bibtex_show = true, domainadaptation = {true} }
2023
-
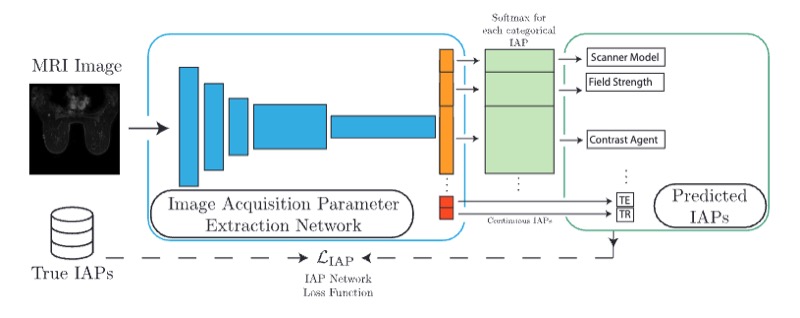 Reverse engineering breast mris: Predicting acquisition parameters directly from imagesNicholas Konz, and Maciej A. MazurowskiMIDL, 2023
Reverse engineering breast mris: Predicting acquisition parameters directly from imagesNicholas Konz, and Maciej A. MazurowskiMIDL, 2023The image acquisition parameters (IAPs) used to create MRI scans are central to defining the appearance of the images. Deep learning models trained on data acquired using certain parameters might not generalize well to images acquired with different parameters. Being able to recover such parameters directly from an image could help determine whether a deep learning model is applicable, and could assist with data harmonization and/or domain adaptation. Here, we introduce a neural network model that can predict many complex IAPs used to generate an MR image with high accuracy solely using the image, with a single forward pass. These predicted parameters include field strength, echo and repetition times, acquisition matrix, scanner model, scan options, and others. Even challenging parameters such as contrast agent type can be predicted with good accuracy. We perform a variety of experiments and analyses of our model’s ability to predict IAPs on many MRI scans of new patients, and demonstrate its usage in a realistic application. Predicting IAPs from the images is an important step toward better understanding the relationship between image appearance and IAPs. This in turn will advance the understanding of many concepts related to the generalizability of neural network models on medical images, including domain shift, domain adaptation, and data harmonization.
@article{konz2023reverse, title = {Reverse engineering breast mris: Predicting acquisition parameters directly from images}, author = {Konz, Nicholas and Mazurowski, Maciej A.}, journal = {MIDL}, year = {2023}, organization = {PMLR}, bibtex_show = true, domainadaptation = {true} } -
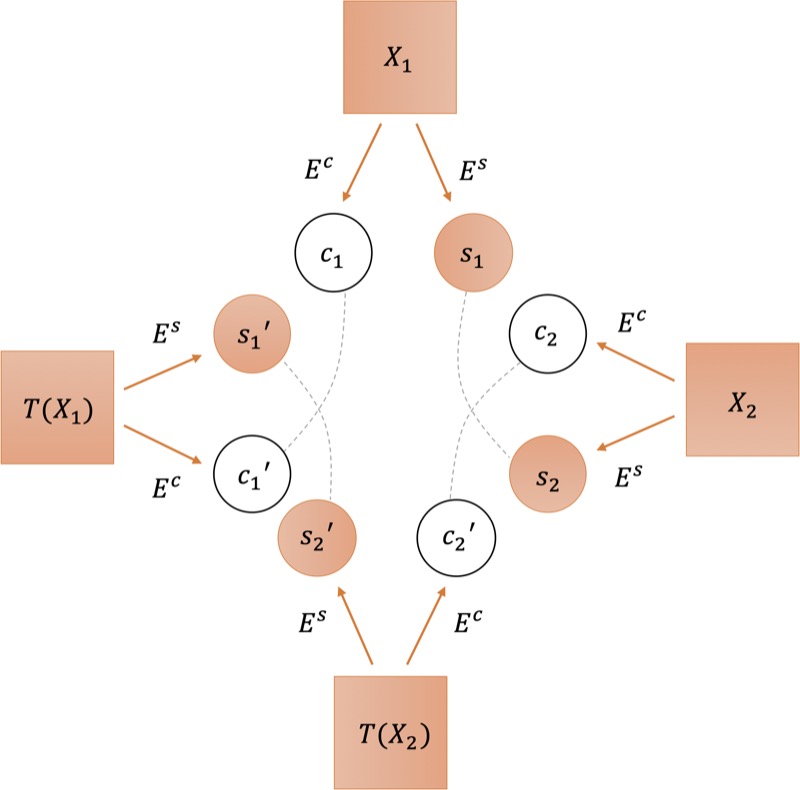 Deep learning for breast mri style transfer with limited training dataShixing Cao, Nicholas Konz, James Duncan, and 1 more authorJournal of Digital imaging, 2023
Deep learning for breast mri style transfer with limited training dataShixing Cao, Nicholas Konz, James Duncan, and 1 more authorJournal of Digital imaging, 2023In this work we introduce a novel medical image style transfer method, StyleMapper, that can transfer medical scans to an unseen style with access to limited training data. This is made possible by training our model on unlimited possibilities of simulated random medical imaging styles on the training set, making our work more computationally efficient when compared with other style transfer methods. Moreover, our method enables arbitrary style transfer: transferring images to styles unseen in training. This is useful for medical imaging, where images are acquired using different protocols and different scanner models, resulting in a variety of styles that data may need to be transferred between. Methods: Our model disentangles image content from style and can modify an image’s style by simply replacing the style encoding with one extracted from a single image of the target style, with no additional optimization required. This also allows the model to distinguish between different styles of images, including among those that were unseen in training. We propose a formal description of the proposed model. Results: Experimental results on breast magnetic resonance images indicate the effectiveness of our method for style transfer. Conclusion: Our style transfer method allows for the alignment of medical images taken with different scanners into a single unified style dataset, allowing for the training of other downstream tasks on such a dataset for tasks such as classification, object detection and others.
@article{cao2023deep, title = {Deep learning for breast mri style transfer with limited training data}, author = {Cao, Shixing and Konz, Nicholas and Duncan, James and Mazurowski, Maciej A}, journal = {Journal of Digital imaging}, volume = {36}, number = {2}, pages = {666--678}, year = {2023}, publisher = {Springer}, bibtex_show = true, domainadaptation = {true} }
Anomaly Detection and Localization
Unsupervised and self-supervised approaches for detecting abnormalities in medical images, with applications ranging from breast cancer screening to general outlier detection in high-resolution imaging data.
2023
-
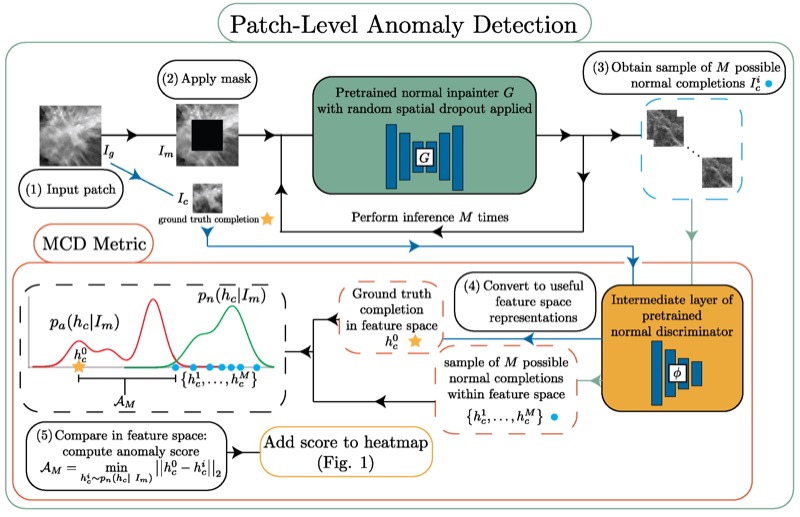 Unsupervised anomaly localization in high-resolution breast scans using deep pluralistic image completionNicholas Konz, Haoyu Dong, and Maciej A. MazurowskiMedical Image Analysis, 2023
Unsupervised anomaly localization in high-resolution breast scans using deep pluralistic image completionNicholas Konz, Haoyu Dong, and Maciej A. MazurowskiMedical Image Analysis, 2023Automated tumor detection in Digital Breast Tomosynthesis (DBT) is a difficult task due to natural tumor rarity, breast tissue variability, and high resolution. Given the scarcity of abnormal images and the abundance of normal images for this problem, an anomaly detection/localization approach could be well-suited. However, most anomaly localization research in machine learning focuses on non-medical datasets, and we find that these methods fall short when adapted to medical imaging datasets. The problem is alleviated when we solve the task from the image completion perspective, in which the presence of anomalies can be indicated by a discrepancy between the original appearance and its auto-completion conditioned on the surroundings. However, there are often many valid normal completions given the same surroundings, especially in the DBT dataset, making this evaluation criterion less precise. To address such an issue, we consider pluralistic image completion by exploring the distribution of possible completions instead of generating fixed predictions. This is achieved through our novel application of spatial dropout on the completion network during inference time only, which requires no additional training cost and is effective at generating diverse completions. We further propose minimum completion distance (MCD), a new metric for detecting anomalies, thanks to these stochastic completions. We provide theoretical as well as empirical support for the superiority over existing methods of using the proposed method for anomaly localization. On the DBT dataset, our model outperforms other state-of-the-art methods by at least 10% AUROC for pixel-level detection.
@article{konz2023picard, title = {Unsupervised anomaly localization in high-resolution breast scans using deep pluralistic image completion}, author = {Konz, Nicholas and Dong, Haoyu and Mazurowski, Maciej A.}, journal = {Medical Image Analysis}, volume = {87}, pages = {102836}, year = {2023}, publisher = {Elsevier}, bibtex_show = true, anomalydetection = {true} } -
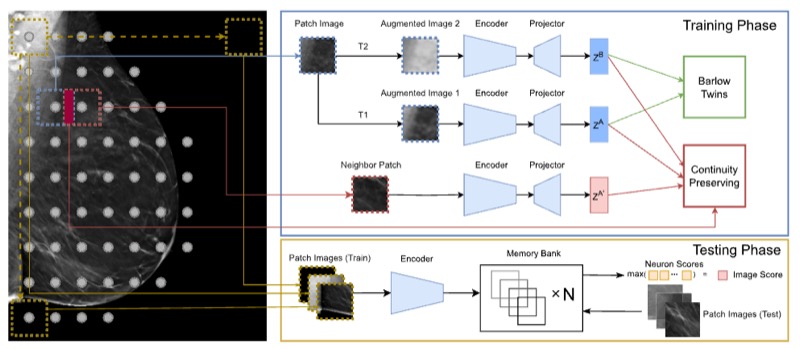 SWSSL: Sliding window-based self-supervised learning for anomaly detection in high-resolution imagesHaoyu Dong, Yifan Zhang, Hanxue Gu, and 3 more authorsIEEE Transactions on Medical Imaging, 2023
SWSSL: Sliding window-based self-supervised learning for anomaly detection in high-resolution imagesHaoyu Dong, Yifan Zhang, Hanxue Gu, and 3 more authorsIEEE Transactions on Medical Imaging, 2023Anomaly detection (AD) aims to determine if an instance has properties different from those seen in normal cases. The success of this technique depends on how well a neural network learns from normal instances. We observe that the learning difficulty scales exponentially with the input resolution, making it infeasible to apply AD to high-resolution images. Resizing them to a lower resolution is a compromising solution and does not align with clinical practice where the diagnosis could depend on image details. In this work, we propose to train the network and perform inference at the patch level, through the sliding window algorithm. This simple operation allows the network to receive high-resolution images but introduces additional training difficulties, including inconsistent image structure and higher variance. We address these concerns by setting the network’s objective to learn augmentation-invariant features. We further study the augmentation function in the context of medical imaging. In particular, we observe that the resizing operation, a key augmentation in general computer vision literature, is detrimental to detection accuracy, and the inverting operation can be beneficial. We also propose a new module that encourages the network to learn from adjacent patches to boost detection performance. Extensive experiments are conducted on breast tomosynthesis and chest X-ray datasets and our method improves 8.03% and 5.66% AUC on image-level classification respectively over the current leading techniques. The experimental results demonstrate the effectiveness of our approach.
@article{dong2023swssl, title = {SWSSL: Sliding window-based self-supervised learning for anomaly detection in high-resolution images}, author = {Dong, Haoyu and Zhang, Yifan and Gu, Hanxue and Konz, Nicholas and Zhang, Yixin and Mazurowski, Maciej A.}, journal = {IEEE Transactions on Medical Imaging}, year = {2023}, publisher = {IEEE}, bibtex_show = true, anomalydetection = {true} }
2021
-
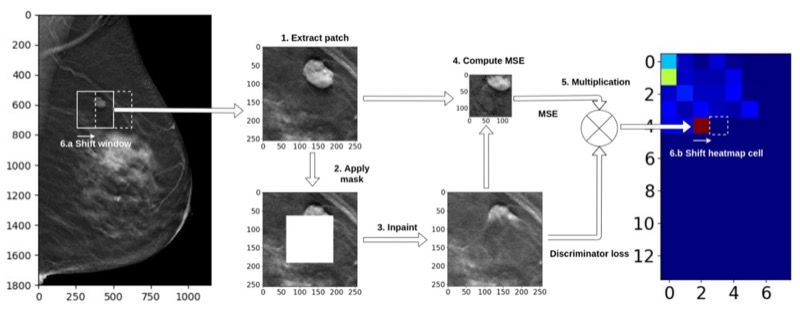 A generative adversarial network-based abnormality detection using only normal images for model training with application to digital breast tomosynthesisAlbert Swiecicki, Nicholas Konz, Mateusz Buda, and 1 more authorScientific reports, 2021
A generative adversarial network-based abnormality detection using only normal images for model training with application to digital breast tomosynthesisAlbert Swiecicki, Nicholas Konz, Mateusz Buda, and 1 more authorScientific reports, 2021Deep learning has shown tremendous potential in the task of object detection in images. However, a common challenge with this task is when only a limited number of images containing the object of interest are available. This is a particular issue in cancer screening, such as digital breast tomosynthesis (DBT), where less than 1% of cases contain cancer. In this study, we propose a method to train an inpainting generative adversarial network to be used for cancer detection using only images that do not contain cancer. During inference, we removed a part of the image and used the network to complete the removed part. A significant error in completing an image part was considered an indication that such location is unexpected and thus abnormal. A large dataset of DBT images used in this study was collected at Duke University. It consisted of 19,230 reconstructed volumes from 4348 patients. Cancerous masses and architectural distortions were marked with bounding boxes by radiologists. Our experiments showed that the locations containing cancer were associated with a notably higher completion error than the non-cancer locations (mean error ratio of 2.77). All data used in this study has been made publicly available by the authors.
@article{swiecicki2021generative, title = {A generative adversarial network-based abnormality detection using only normal images for model training with application to digital breast tomosynthesis}, author = {Swiecicki, Albert and Konz, Nicholas and Buda, Mateusz and Mazurowski, Maciej A}, journal = {Scientific reports}, volume = {11}, number = {1}, pages = {10276}, year = {2021}, publisher = {Nature Publishing Group UK London}, bibtex_show = true, anomalydetection = {true} }
2018
-
 Robust chauvenet outlier rejectionMP Maples, DE Reichart, Nicholas Konz, and 7 more authorsThe Astrophysical Journal Supplement Series, 2018
Robust chauvenet outlier rejectionMP Maples, DE Reichart, Nicholas Konz, and 7 more authorsThe Astrophysical Journal Supplement Series, 2018Sigma clipping is commonly used in astronomy for outlier rejection, but the number of standard deviations beyond which one should clip data from a sample ultimately depends on the size of the sample. Chauvenet rejection is one of the oldest, and simplest, ways to account for this, but, like sigma clipping, depends on the sample’s mean and standard deviation, neither of which are robust quantities: Both are easily contaminated by the very outliers they are being used to reject. Many, more robust measures of central tendency, and of sample deviation, exist, but each has a tradeoff with precision. Here, we demonstrate that outlier rejection can be both very robust and very precise if decreasingly robust but increasingly precise techniques are applied in sequence. To this end, we present a variation on Chauvenet rejection that we call "robust" Chauvenet rejection (RCR), which uses three decreasingly robust/increasingly precise measures of central tendency, and four decreasingly robust/increasingly precise measures of sample deviation. We show this sequential approach to be very effective for a wide variety of contaminant types, even when a significant – even dominant – fraction of the sample is contaminated, and especially when the contaminants are strong. Furthermore, we have developed a bulk-rejection variant, to significantly decrease computing times, and RCR can be applied both to weighted data, and when fitting parameterized models to data. We present aperture photometry in a contaminated, crowded field as an example. RCR may be used by anyone at https://github.com/nickk124/robust-outlier-rejection, and source code is available there as well.
@article{maples2018robust, title = {Robust chauvenet outlier rejection}, author = {Maples, MP and Reichart, DE and Konz, Nicholas and Berger, TA and Trotter, AS and Martin, JR and Dutton, DA and Paggen, ML and Joyner, RE and Salemi, CP}, journal = {The Astrophysical Journal Supplement Series}, volume = {238}, number = {1}, pages = {2}, year = {2018}, publisher = {IOP Publishing}, bibtex_show = true, anomalydetection = {true} }
Misc. Breast Imaging Analysis
Specialized methods for breast imaging applications, including lesion detection algorithms, registration techniques, and style transfer methods tailored to the unique challenges of breast MRI and tomosynthesis.
2025
-
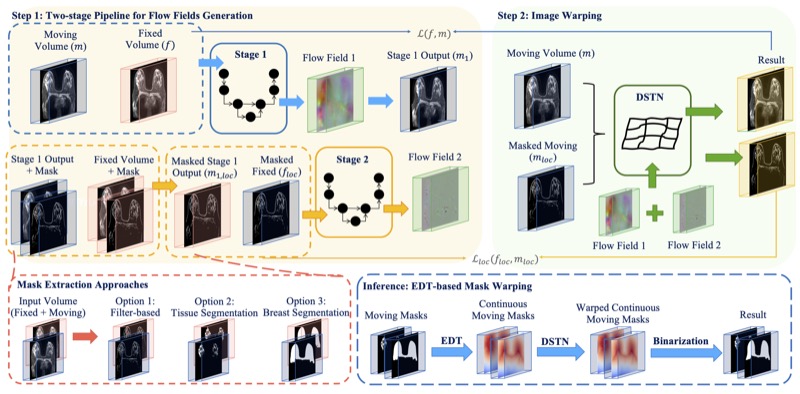 GuidedMorph: Two-Stage Deformable Registration for Breast MRIYaqian Chen, Hanxue Gu, Haoyu Dong, and 5 more authorsarXiv preprint, 2025
GuidedMorph: Two-Stage Deformable Registration for Breast MRIYaqian Chen, Hanxue Gu, Haoyu Dong, and 5 more authorsarXiv preprint, 2025Accurately registering breast MR images from different time points enables the alignment of anatomical structures and tracking of tumor progression, supporting more effective breast cancer detection, diagnosis, and treatment planning. However, the complexity of dense tissue and its highly non-rigid nature pose challenges for conventional registration methods, which primarily focus on aligning general structures while overlooking intricate internal details. To address this, we propose GuidedMorph, a novel two-stage registration framework designed to better align dense tissue. In addition to a single-scale network for global structure alignment, we introduce a framework that utilizes dense tissue information to track breast movement. The learned transformation fields are fused by introducing the Dual Spatial Transformer Network (DSTN), improving overall alignment accuracy. A novel warping method based on the Euclidean distance transform (EDT) is also proposed to accurately warp the registered dense tissue and breast masks, preserving fine structural details during deformation. The framework supports paradigms that require external segmentation models and with image data only. It also operates effectively with the VoxelMorph and TransMorph backbones, offering a versatile solution for breast registration. We validate our method on ISPY2 and internal dataset, demonstrating superior performance in dense tissue, overall breast alignment, and breast structural similarity index measure (SSIM), with notable improvements by over 13.01% in dense tissue Dice, 3.13% in breast Dice, and 1.21% in breast SSIM compared to the best learning-based baseline.
@article{chen2025guidedmorph, title = {GuidedMorph: Two-Stage Deformable Registration for Breast MRI}, author = {Chen, Yaqian and Gu, Hanxue and Dong, Haoyu and Li, Qihang and Chen, Yuwen and Konz, Nicholas and Li, Lin and Mazurowski, Maciej A}, journal = {arXiv preprint}, year = {2025}, bibtex_show = true, breastimaging = {true} }
2023
-
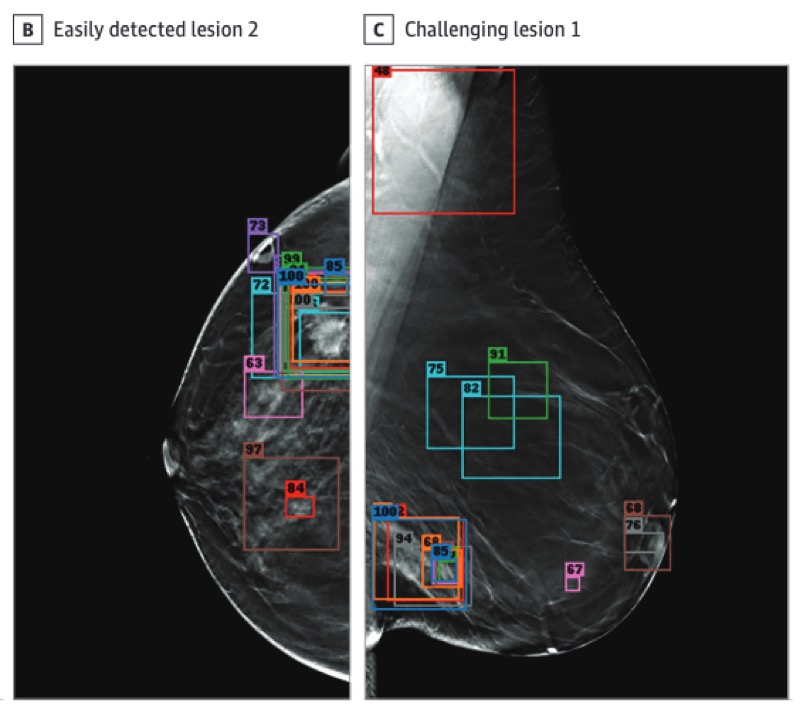 A competition, benchmark, code, and data for using artificial intelligence to detect lesions in digital breast tomosynthesisNicholas Konz, Mateusz Buda, Hanxue Gu, and 8 more authorsJAMA Network Open, 2023
A competition, benchmark, code, and data for using artificial intelligence to detect lesions in digital breast tomosynthesisNicholas Konz, Mateusz Buda, Hanxue Gu, and 8 more authorsJAMA Network Open, 2023Importance: An accurate and robust artificial intelligence (AI) algorithm for detecting cancer in digital breast tomosynthesis (DBT) could significantly improve detection accuracy and reduce health care costs worldwide. Objectives: To make training and evaluation data for the development of AI algorithms for DBT analysis available, to develop well-defined benchmarks, and to create publicly available code for existing methods. Design, Setting, and Participants: This diagnostic study is based on a multi-institutional international grand challenge in which research teams developed algorithms to detect lesions in DBT. A data set of 22 032 reconstructed DBT volumes was made available to research teams. Phase 1, in which teams were provided 700 scans from the training set, 120 from the validation set, and 180 from the test set, took place from December 2020 to January 2021, and phase 2, in which teams were given the full data set, took place from May to July 2021. Main Outcomes and Measures: The overall performance was evaluated by mean sensitivity for biopsied lesions using only DBT volumes with biopsied lesions; ties were broken by including all DBT volumes. Results: A total of 8 teams participated in the challenge. The team with the highest mean sensitivity for biopsied lesions was the NYU B-Team, with 0.957 (95% CI, 0.924-0.984), and the second-place team, ZeDuS, had a mean sensitivity of 0.926 (95% CI, 0.881-0.964). When the results were aggregated, the mean sensitivity for all submitted algorithms was 0.879; for only those who participated in phase 2, it was 0.926. Conclusions and Relevance: In this diagnostic study, an international competition produced algorithms with high sensitivity for using AI to detect lesions on DBT images. A standardized performance benchmark for the detection task using publicly available clinical imaging data was released, with detailed descriptions and analyses of submitted algorithms accompanied by a public release of their predictions and code for selected methods. These resources will serve as a foundation for future research on computer-assisted diagnosis methods for DBT, significantly lowering the barrier of entry for new researchers.
@article{konz2023competition, title = {A competition, benchmark, code, and data for using artificial intelligence to detect lesions in digital breast tomosynthesis}, author = {Konz, Nicholas and Buda, Mateusz and Gu, Hanxue and Saha, Ashirbani and Yang, Jichen and Ch{\l}{\k{e}}dowski, Jakub and Park, Jungkyu and Witowski, Jan and Geras, Krzysztof J and Shoshan, Yoel and others}, journal = {JAMA Network Open}, volume = {6}, number = {2}, pages = {e230524--e230524}, year = {2023}, publisher = {American Medical Association}, bibtex_show = true, breastimaging = {true} }
2022
-
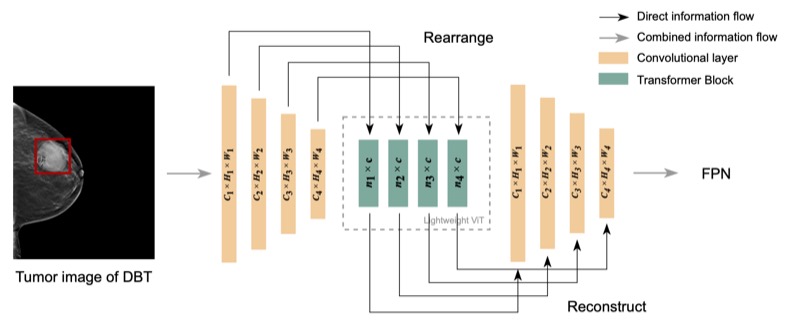 Lightweight transformer backbone for medical object detectionYifan Zhang, Haoyu Dong, Nicholas Konz, and 2 more authors2022
Lightweight transformer backbone for medical object detectionYifan Zhang, Haoyu Dong, Nicholas Konz, and 2 more authors2022Lesion detection in digital breast tomosynthesis (DBT) is an important and a challenging problem characterized by a low prevalence of images containing tumors. Due to the label scarcity problem, large deep learning models and computationally intensive algorithms are likely to fail when applied to this task. In this paper, we present a practical yet lightweight backbone to improve the accuracy of tumor detection. Specifically, we propose a novel modification of visual transformer (ViT) on image feature patches to connect the feature patches of a tumor with healthy backgrounds of breast images and form a more robust backbone for tumor detection. To the best of our knowledge, our model is the first work of Transformer backbone object detection for medical imaging. Our experiments show that this model can considerably improve the accuracy of lesion detection and reduce the amount of labeled data required in typical ViT. We further show that with additional augmented tumor data, our model significantly outperforms the Faster R-CNN model and state-of-the-art SWIN transformer model.
@article{zhang2022lightweight, title = {Lightweight transformer backbone for medical object detection}, author = {Zhang, Yifan and Dong, Haoyu and Konz, Nicholas and Gu, Hanxue and Mazurowski, Maciej A}, booktitle = {CaPTion @ MICCAI}, pages = {47--56}, year = {2022}, organization = {Springer}, bibtex_show = true, breastimaging = {true} }